Back to research page

Back to top
Highlights
- Optimized PatchMatch for Accurate Label fusion (OPAL) and segmentation of 3D MRI structures
- New multi-scale and multi-feature framework
- Fast and accurate results on hippocampus segmentation (<1s)
- Inter-class expert reliability is reached on healthy and pathological datasets
- Extensions to Cerebellum segmentation and Alzheimer's disease classification
- Integration into the online volBrain platform
- Use of a linearly registered database with expert-based segmentation of structures
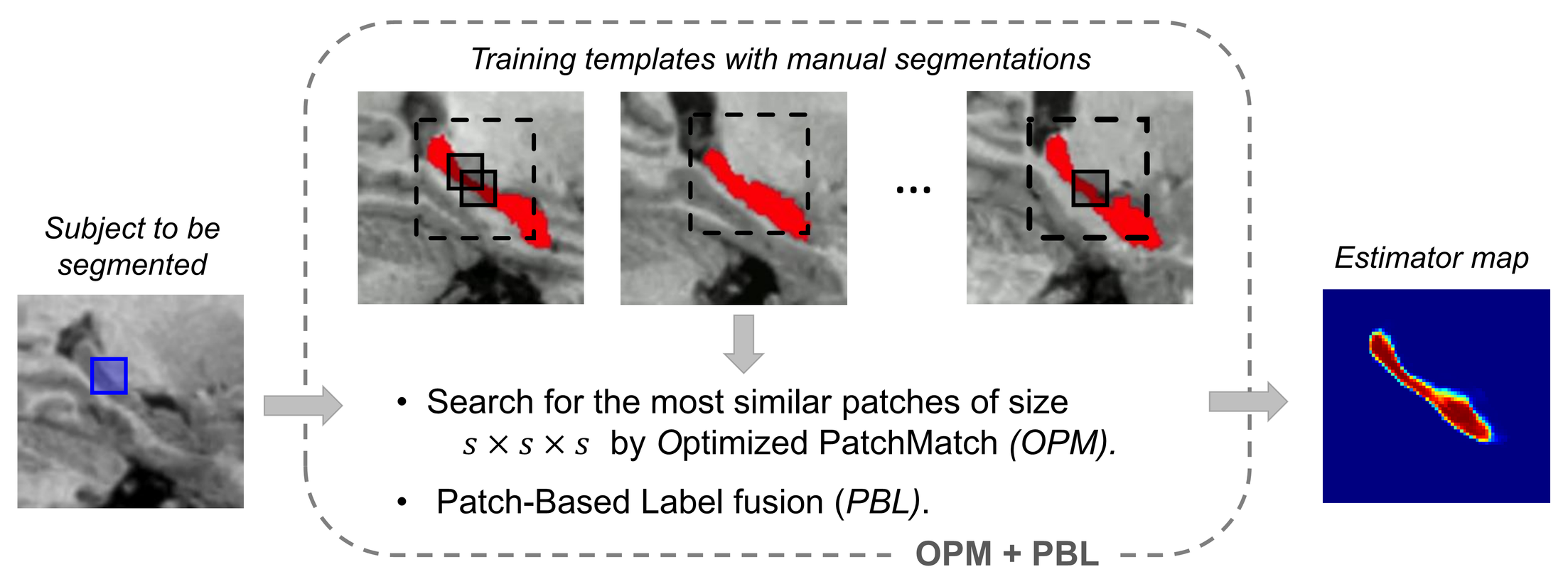
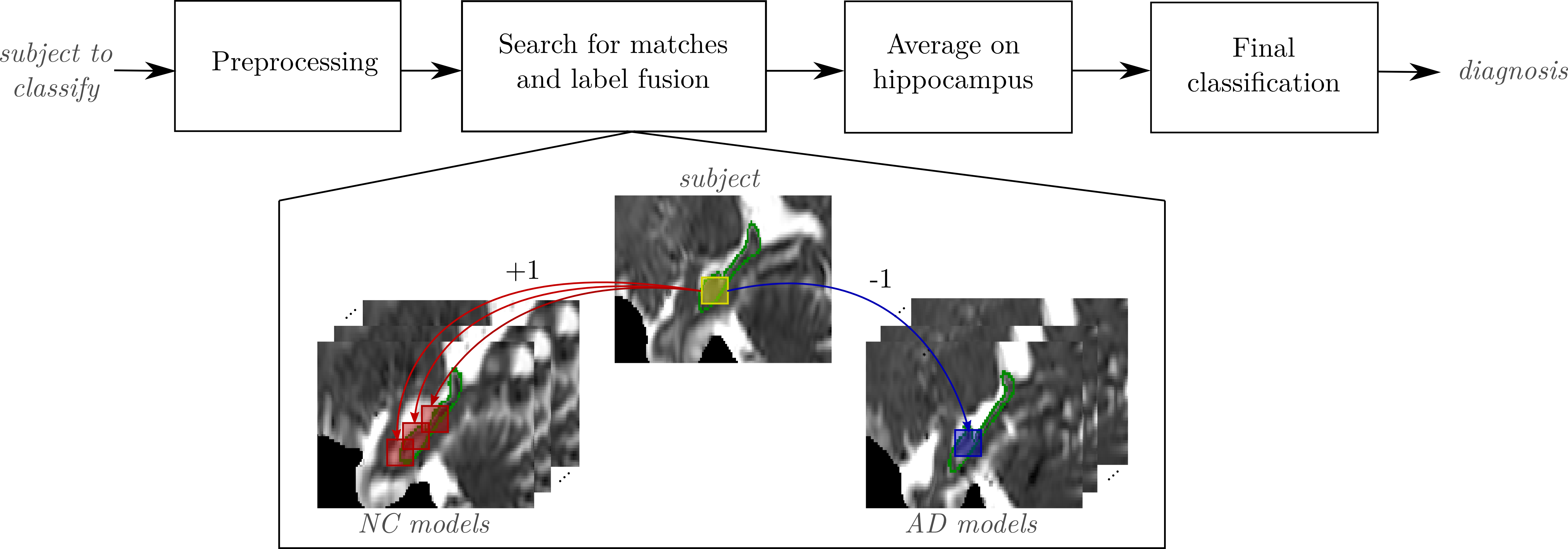
- Patch-based approximate nearest neighbors (ANN) matching and transfer of manual segmentation information
Fast search of patch-based neighbors
- PatchMatch algorithm [6] introduced for 2D patch matching between 2 images
- New Optimized PatchMatch (OPM) algorithm for multiple 3D patch matching within a database
- Search based on propagation step (PS) of good matches, with new constrained initialization (CI) and random search (CRS) steps

Label fusion from expert-based segmentations
- Patch-wise label fusion from selected ANN, inspired from non-local means [7]

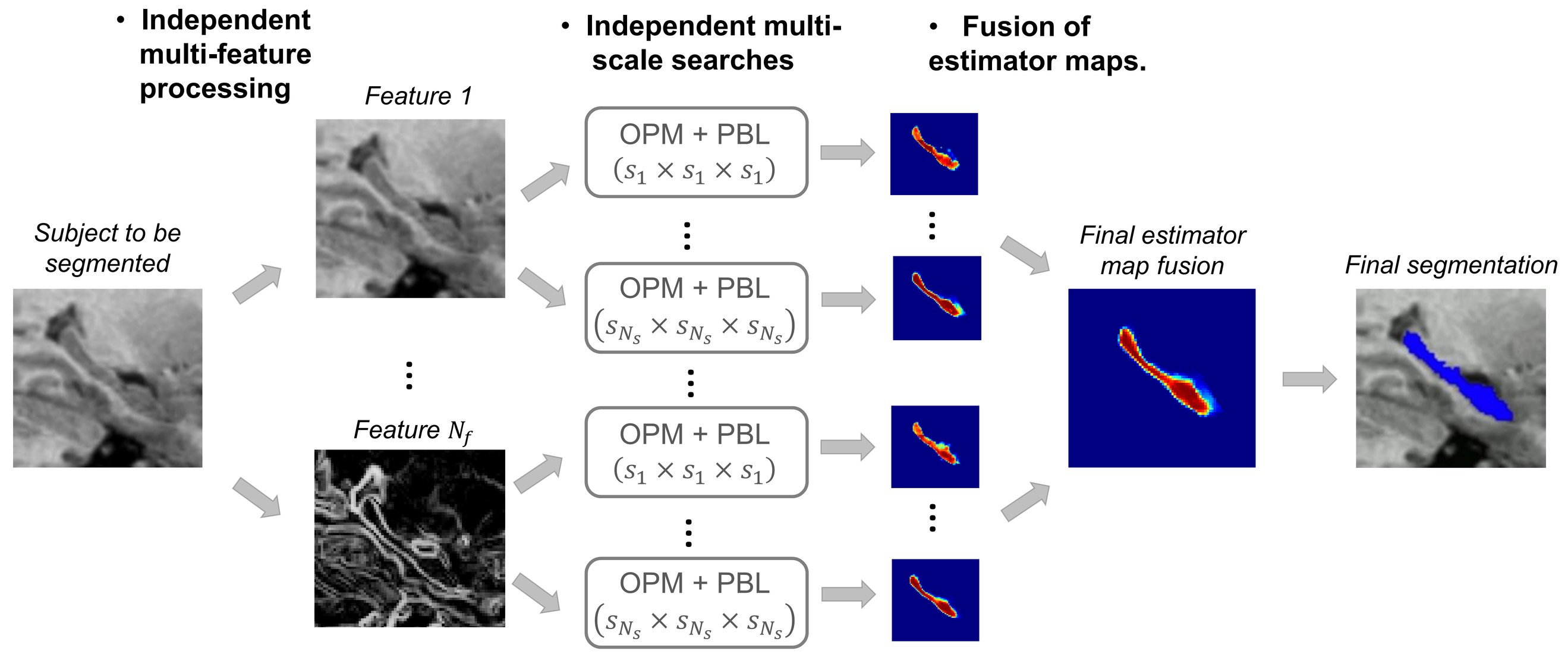
Multi-scale and multi-feature framework
- Fast ANN search and label fusion on several scales and features
- Late fusion of estimator maps

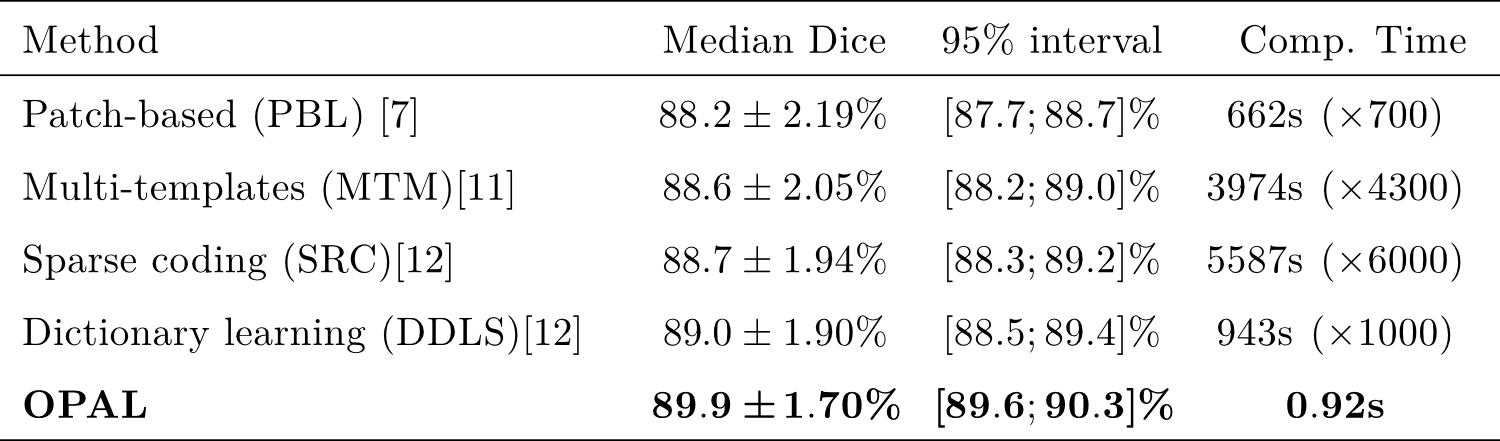
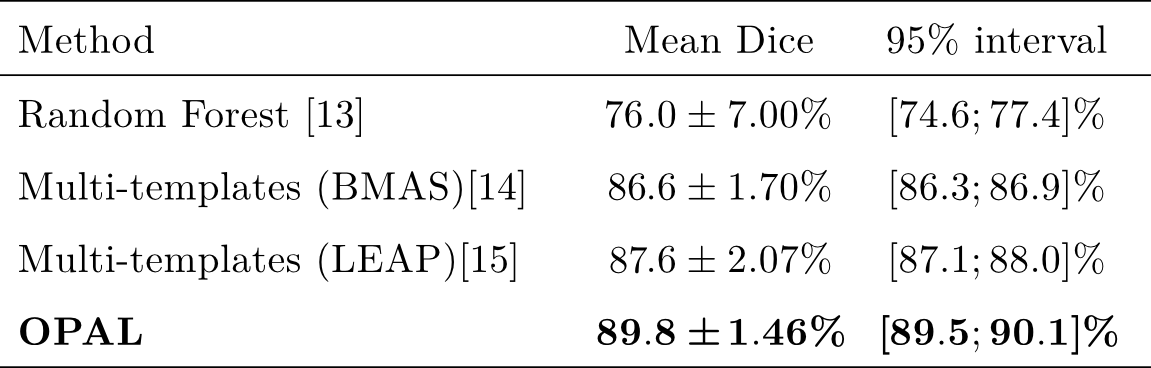
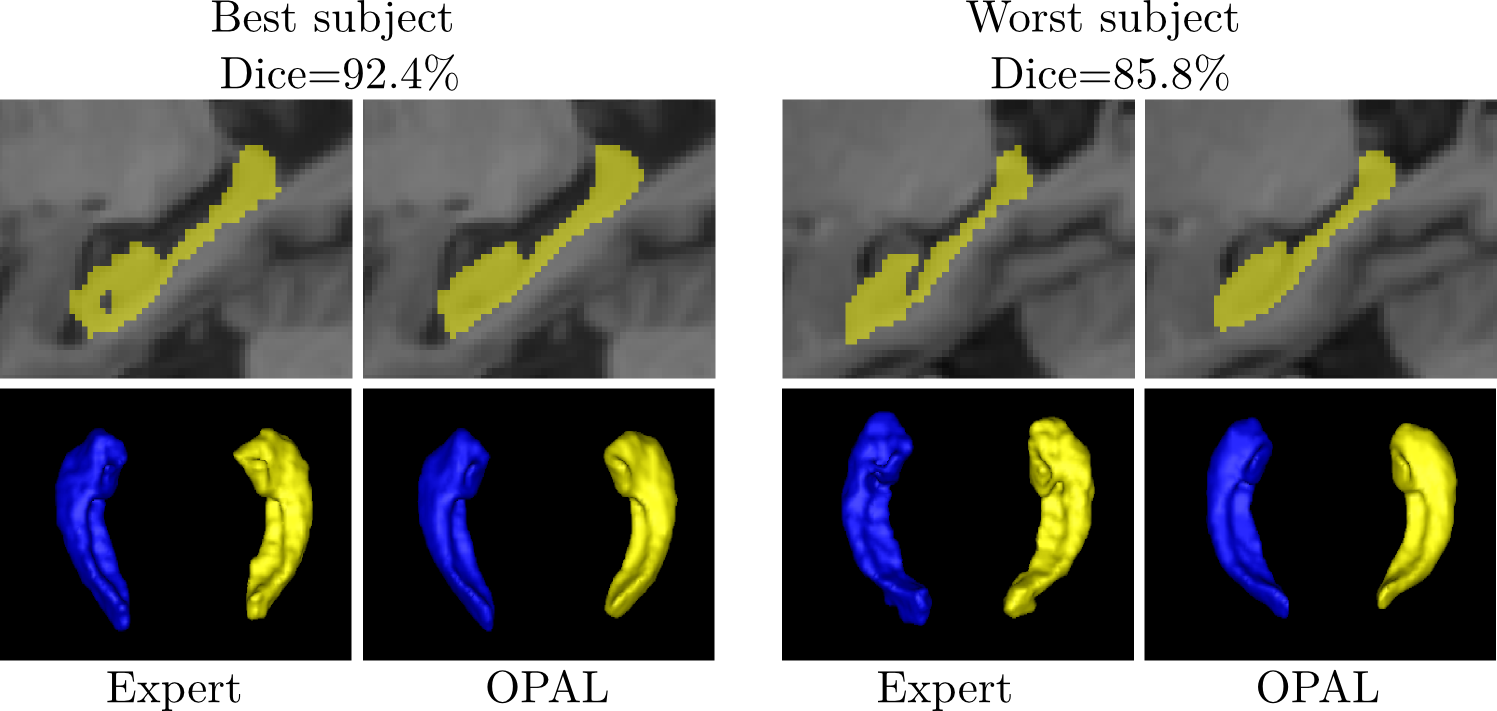
- Leave-one-out validation procedure. Comparison on the Dice index [8]
- OPAL is more accurate and much faster than state-of-the-art methods
Main (to cite)
- R. Giraud, V.-T. Ta, N. Papadakis, et al.: An Optimized PatchMatch for multi-scale and multi-feature label fusion
NeuroImage, 2015

Others
- V.-T. Ta, R. Giraud, D. L. Collins and P. Coupé: Optimized PatchMatch for Near Real Time and Accurate Label Fusion
Int. Conf. on Medical Image Computing and Computer Assisted Interventions (MICCAI), 2014

- R. Giraud, V.-T. Ta, et al.: Optimisation de l'algorithme PatchMatch pour la segmentation de structures anatomiques
Groupement de Recherche en Traitement du Signal et des Images (GRETSI), 2015


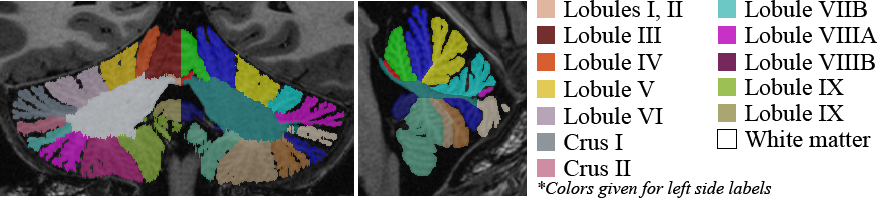
- New method for cerebellum lobule segmentation on 3D MRI
- Extension of OPAL to multiple structure segmentation
- Integrated into online platform volBrain

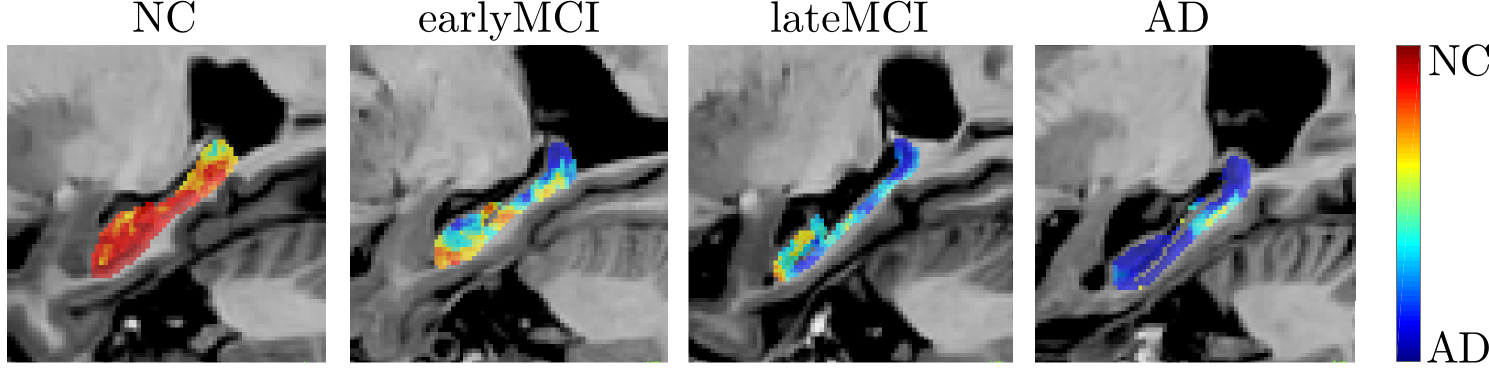
- Fast feature extraction on DTI (Diffusion Tensor Imaging) using OPAL
- The extracted features lead to more accurate Alzheimer's disease detection
(AD = Alzheimer's Disease, NC = Normal Control, MCI = Mild Cognitive Impairment)


- C. Barnes, E. Shechtman, A. Finkelstein, et al.: PatchMatch: A randomized correspondence algorithm for structural image editing
ACM Transactions of Graphics, 2009
- P. Coupé, J. V. Manjón, et al.: Patch-based segmentation using expert priors: application to hippocampus and ventricle segmentation
NeuroImage, 2011
- A. P. Zijdenbos, B. M. Dawant, R. A. Margolin, et al.: Morphometric analysis of white matter lesions in MR images: method and
validation
IEEE Trans. on Medical Imaging, 1994
- J. C. Mazziotta, A. W. Toga, A. C. Evans, et al.: A probabilistic atlas of the human brain: theory and rationale for its development
NeuroImage, 1995
- M. Boccardi, et al.: Delphi definition of the EADC-ADNI harmonized protocol for hippocampal segmentation on magnetic resonance
Alzheimer's & Dementia, 2014
- D. L. Collins and J. C. Pruessner: Towards accurate, automatic segmentation of the hippocampus and amygdala from MRI by augmenting ANIMAL
with a template library and label fusion.
NeuroImage, 2010
- T. Tong, R. Wolz, P. Coupé, J. V. Hajnal, D. Rueckert, et al.: Segmentation of MR images via discriminative dictionary learning and sparse coding: Application to hippocampus labeling.
NeuroImage, 2013
- S. Tangaro, N. Amoroso, et al.: Automated voxel-by-voxel tissue classification for hippocampal segmentation: Methods and validation
Physica Medica, 2014
- F. Roche, J. Schaerer, S. Gouttard, et al.: Accuracy of BMAS Hippocampus Segmentation Using the
Harmonized Hippocampal Protocol
Alzheimer's & Dementia, 2014
- K. R. Gray, M. Austin, et al.: Integration of EADC-ADNI Harmonised hippocampus labels into the LEAP automated segmentation technique
Alzheimer's & Dementia, 2014
Back to top